


Next: 12.6 Low-Dispersion Resampled Image
Up: 12 Final Archive Data
Previous: 12.4 Linearized Image FITS
The VD defines the final SI coordinate values in the x (wavelength)
and y (spatial) directions for every LI pixel. The final coordinates
in SI space for any photometrically-corrected pixel in the LILO/LIHI are
determined by:
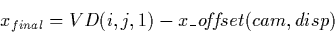
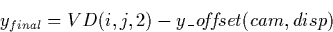
where i and j range from 1 to 768, and
x_offset and y_offset
are given in the following table.
| |
x_offset |
y_offset |
| |
disp=L |
H |
L |
H |
| LWP |
100 |
|
297 |
|
| LWR |
100 |
|
250 |
|
| SWP |
130 |
|
490 |
|
The output displacements between the SI and LI coordinates are
recoverable by:
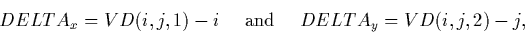
where i and j range from 1 to 768. xfinal and yfinal
contain the final x and y coordinates in the SILO/SIHI. The x and y
coordinates of the displacement vectors are stored as a 3-D primary
array consisting of 768x768x2 elements. The displacements are coded as
32-bit, floating point numbers.
The XC allow the user to recover the calculated displacement vectors,
mapping the science image (in raw space) to the ITF. For each of the
approximately 500 (140 for low dispersion) points used to obtain the
displacement between the science image and the corresponding level of
the ITF, the binary table extension will contain the following columns
of information: science image x-position (I*2), science image
y-position (I*2), ITF x-position at position of best match (R*4),
ITF y-position at position of best match (R*4), the cross-correlation
coefficient (R*4), number of points used to calculate the coefficient
(I*2), and the ITF level used in the correlation (I*2). The x and y
positions correspond to the sample and line numbers in the RI. The
resulting ITF positions of the best match are pre-filtered positions
(before invalid matches have been identified and deleted) and will not
necessarily correspond exactly to the photometric registration
displacement components utilized to create the final displacement
vector.
Basic keywords in the VDLO/VDHI headers and binary table extensions
are shown in Table 12.7. Note that the CTYPE1 and CTYPE3 keyword
values listed and as stored in the archived VDLO/VDHI are incorrect and
should be interchanged. Unfortunately, this error was not discovered
until the majority of images were processed and so was left uncorrected
for consistency. Note also that the VDLO/VDHI will not be available for
images processed at VILSPA, nor for images processed at GSFC after
July 31, 1997.
Table 12.7:
VDLO/VDHI - Basic FITS Keywords
| Keyword and value |
Description |
|---|
SIMPLE = T |
Standard FITS Format |
BITPIX = -32 |
IEEE single precision floating
point |
NAXIS = 3 |
Three-dimensional image |
NAXIS1 = 768 |
Dimension along x-axis |
NAXIS2 = 768 |
Dimension along y-axis |
NAXIS3 = 2 |
Dimension along z-axis |
EXTEND = T |
Extensions are present |
CTYPE1 = ' ' |
Units x-axis |
CTYPE2 = 'PIXEL ' |
Units y-axis |
CTYPE3 = 'PIXEL ' |
Units z-axis |
BUNIT = 'PIXEL ' |
Pixel units |
TELESCOP= 'IUE ' |
International Ultraviolet
Explorer |
FILENAME= 'AAAnnnnn.VDdd' |
Filename(camera)(number).VD(disp) |
DATE = 'dd/mm/yy' |
Date file was written |
ORIGIN = 'VILSPA ' |
Institution generating the file |
DATAMIN = nnnnn.n |
Minimum pixel value |
DATAMAX = nnnnn.n |
Maximum pixel value |
XTENSION= 'BINTABLE' |
Table extension |
BITPIX = 8 |
Binary data |
NAXIS = 2 |
Two-dimensional table array |
NAXIS1 = 20 |
Width of table in bytes |
NAXIS2 = nnn |
Number of entries in table |
PCOUNT = 0 |
Number of bytes following data
matrix |
GCOUNT = 1 |
Number of groups |
TFIELDS = 7 |
Number of fields in each row |
TFORM1 = '1I ' |
Count and data type for field 1 |
TTYPE1 = 'XRAW ' |
Science image x-position |
TUNIT1 = 'PIXEL ' |
Unit is pixels |
TFORM2 = '1I ' |
Count and data type for field 2 |
TTYPE2 = 'YRAW ' |
Science image y-position |
TUNIT2 = 'PIXEL ' |
Unit is pixel |
TFORM3 = '1E ' |
Count and data type for field 3 |
TTYPE3 = 'XITF ' |
ITF x-position of best match |
TUNIT3 = 'PIXEL ' |
Unit is pixel |
TFORM4 = '1E ' |
Count and data type for field 4 |
TTYPE4 = 'YITF ' |
ITF y-position of best match |
TUNIT4 = 'PIXEL ' |
Unit is pixel |
TFORM5 = '1E ' |
Count and data type for field 5 |
TTYPE5 = 'XCOEFF ' |
Cross correlation coefficient |
TUNIT5 = ' ' |
Unitless |
TFORM6 = '1I ' |
Count and data type for field 6 |
TTYPE6 = 'NPOINTS ' |
Number of points used |
TUNIT6 = ' ' |
Unitless |
TFORM7 = '1I ' |
Count and data type for field 7 |
TTYPE7 = 'ITFLEVEL' |
ITF level |
TUNIT7 = ' ' |
Unitless |
FILENAME= 'AAAnnnnn.XCdd' |
Filename (camera)(number).XC(disp) |
EXTNAME = 'XCOEFF ' |
Cross correlation coefficients |



Next: 12.6 Low-Dispersion Resampled Image
Up: 12 Final Archive Data
Previous: 12.4 Linearized Image FITS
Karen Levay
12/4/1997
![]()
![]()
![]()